The ultimate aim of our lab is to understand how genome makes its code work. Our focus is on two main themes.
Evolutionary Genomics
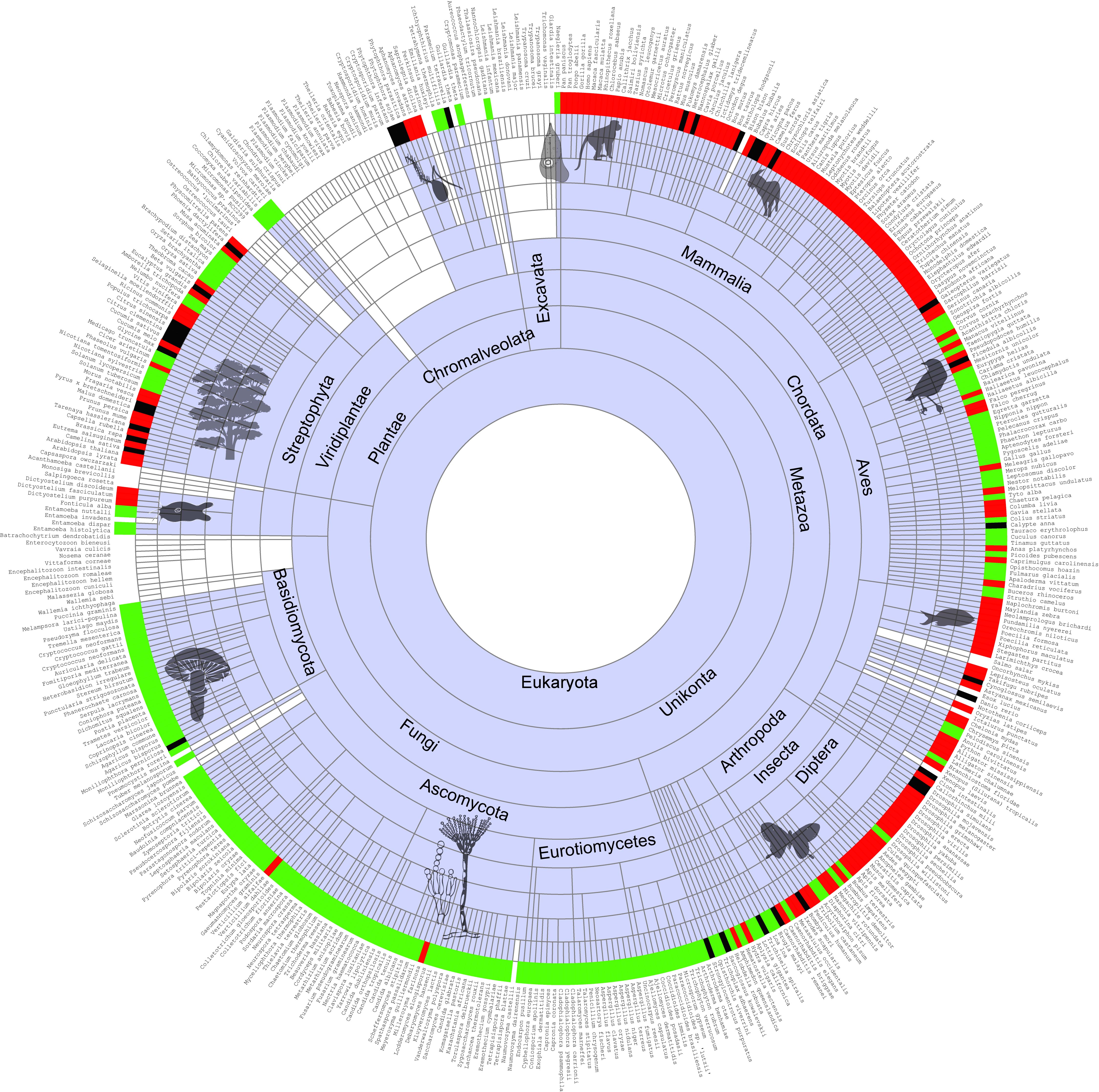
The vast amount of genomic datasets get harder to analyze day by day. Computational tool developers have been trying to catch up with the Next Generation Sequencing data accumulation. However, there is also a need to apply a biologist perspective to individual cases in order to achieve a precision we would need in molecular medicine. We apply computational methods and develop approaches to understand how proteins work. We are primarily interested in determining a protein’s function and the importance of each amino acid. We benefit from molecular evolution methods.
Eukaryotic Cell Signaling
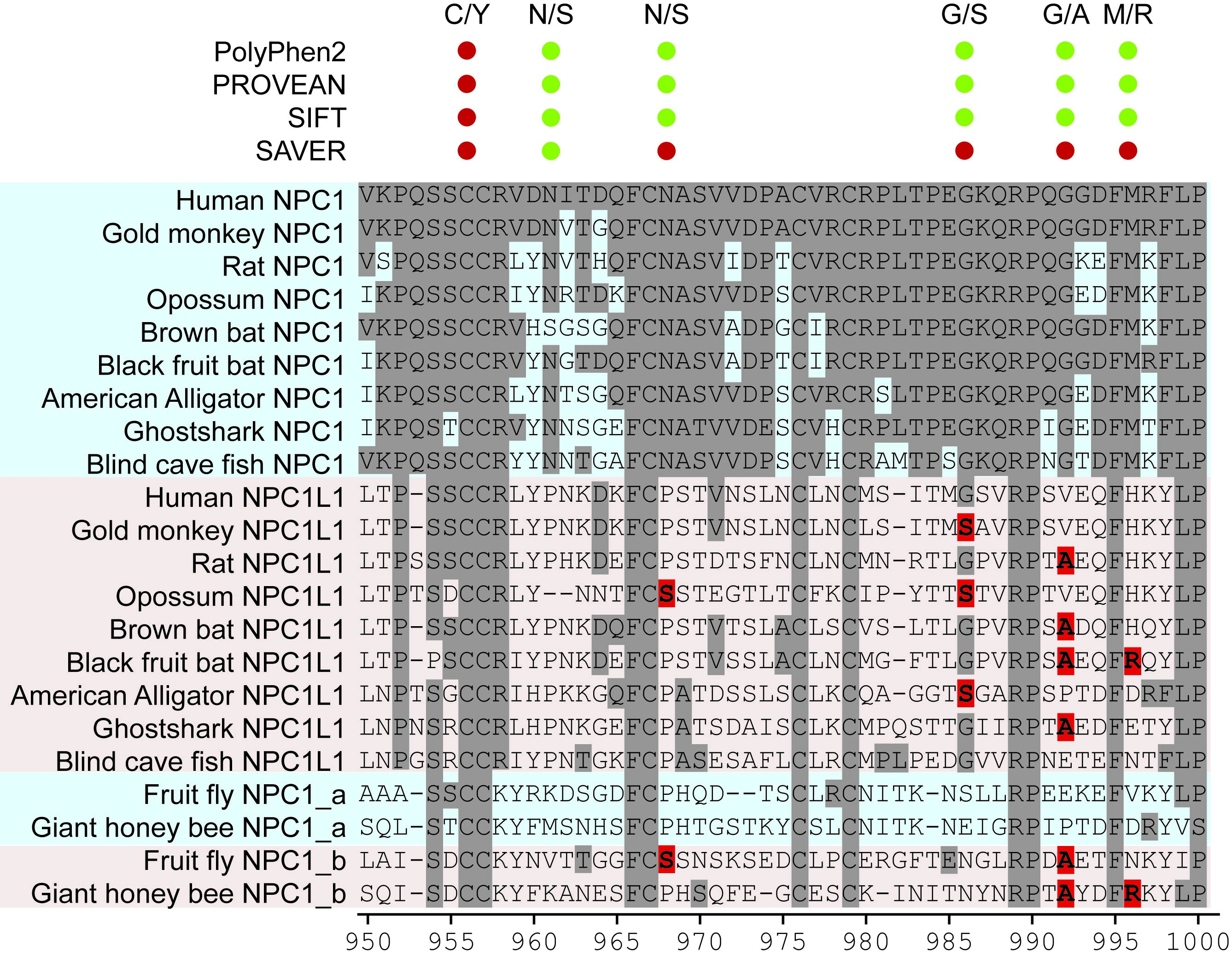
We see, hear and feel; therefore we are! How about our cells? They feel the environmental changes with proteins called receptors and initiate molecular signaling processes. We are particularly interested in a eukaryotic receptor family, G protein-coupled receptors (GPCRs). GPCRs are therapeutically important as they are targeted by more than 1/3 of the marketed drugs. The GPCR evolution has yet to be identified precisely. If we succeed to understand the complex evolution of this receptor family we will have a better understanding of the function and importance of each residue of each subfamily.
Rare Mendelian Disorders
The genetic disorders that affect a small fraction of a population are defined “rare”. However, the number of rare disorders is so high that 1 in every 17 people will be affected by a rare genetic disorder. Therefore rare disorders should not be neglected. The genetic diagnosis of a spectrum of rare diseases has been a major problem. The prediction performance of the automated algorithms is far from the desired level. In our lab, we will attempt to study the evolution of the genes associated with Mendelian diseases and develop computational tools in predicting the effect of amino acid substitutions on protein function.
Here is the detalied information of how we address this problem.
DNA Damage and Repair, Circadian Clock Regulation
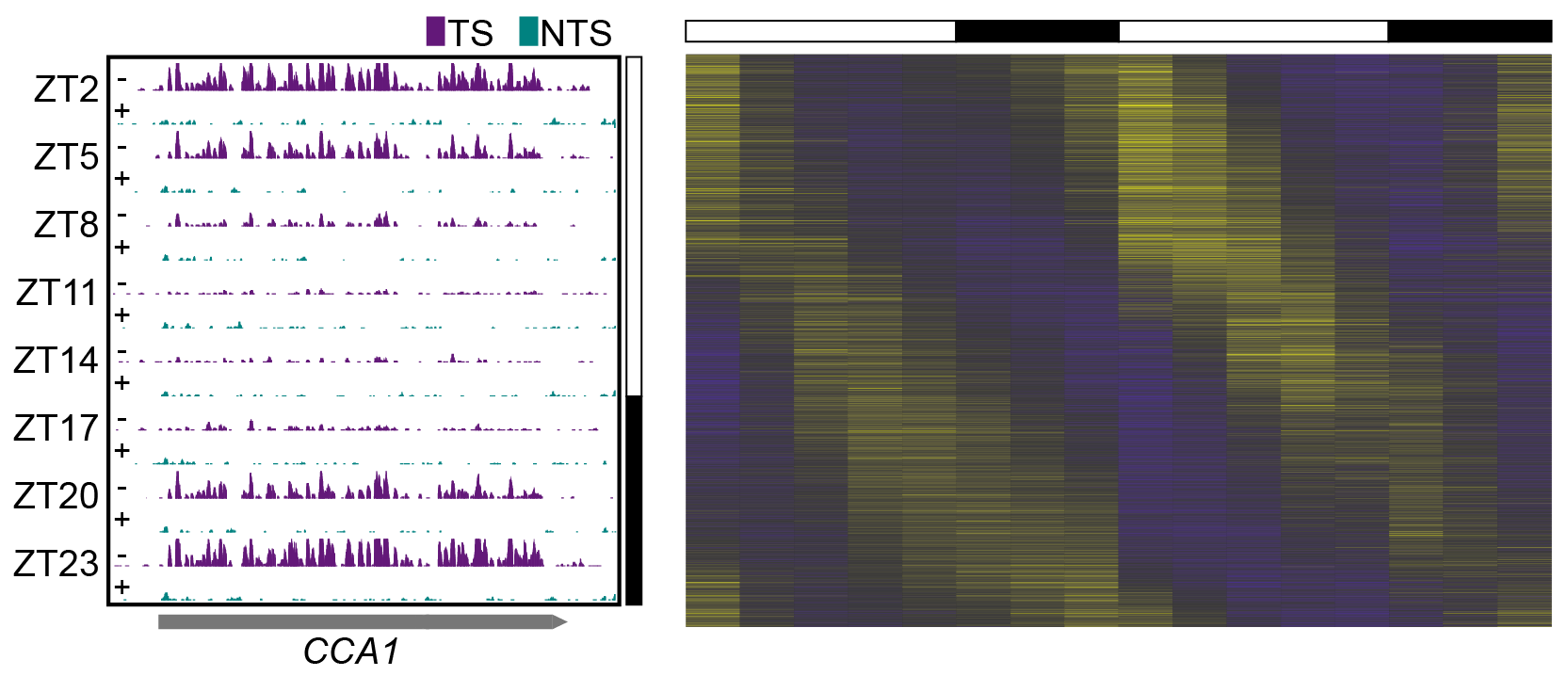
How is our genome damaged? How do our cells cope with the damage? Fortunately, we and other organisms have a cellular mechanism, DNA repair, to remove the DNA damage induced by internal and external factors. We are interested in understanding the dynamics of DNA damage and repair events in genomes. Recently, a number of genome-wide sequencing technologies have been developed to map damage and repair events onto genomes of interest. We apply bioinformatics applications to reveal the genomic factors playing role in DNA damage and repair bias. We actively collaborate with Aziz Sancar Lab and analyze genomic damage and repair profiles under a variety of conditions.
Lab Tools and Resources
We develop tools and resources to enable biologists and bioinformaticians to benefit from our studies. Please visit tools for details.
Funding
- Sabanci University
- Personal Research Funds (yearly)
- Starting Grant (2018-2019)
- EMBO
- Installation Grant (2019-2024)
- Science Academy, Turkey
- BAGEP Young Scientist Grant (2019-2021)
- TUBITAK
- International Fellowship for Outstanding Researchers Programme (2020-2023)
- Return Fellowship Programme (2018-2019)
- TUSEB
- Systems Biology and Bioinformatics Strategic R&D (2020-2022)




