PUBLICATIONS
CDvist: a webserver for identification and visualization of conserved domains in protein sequences.
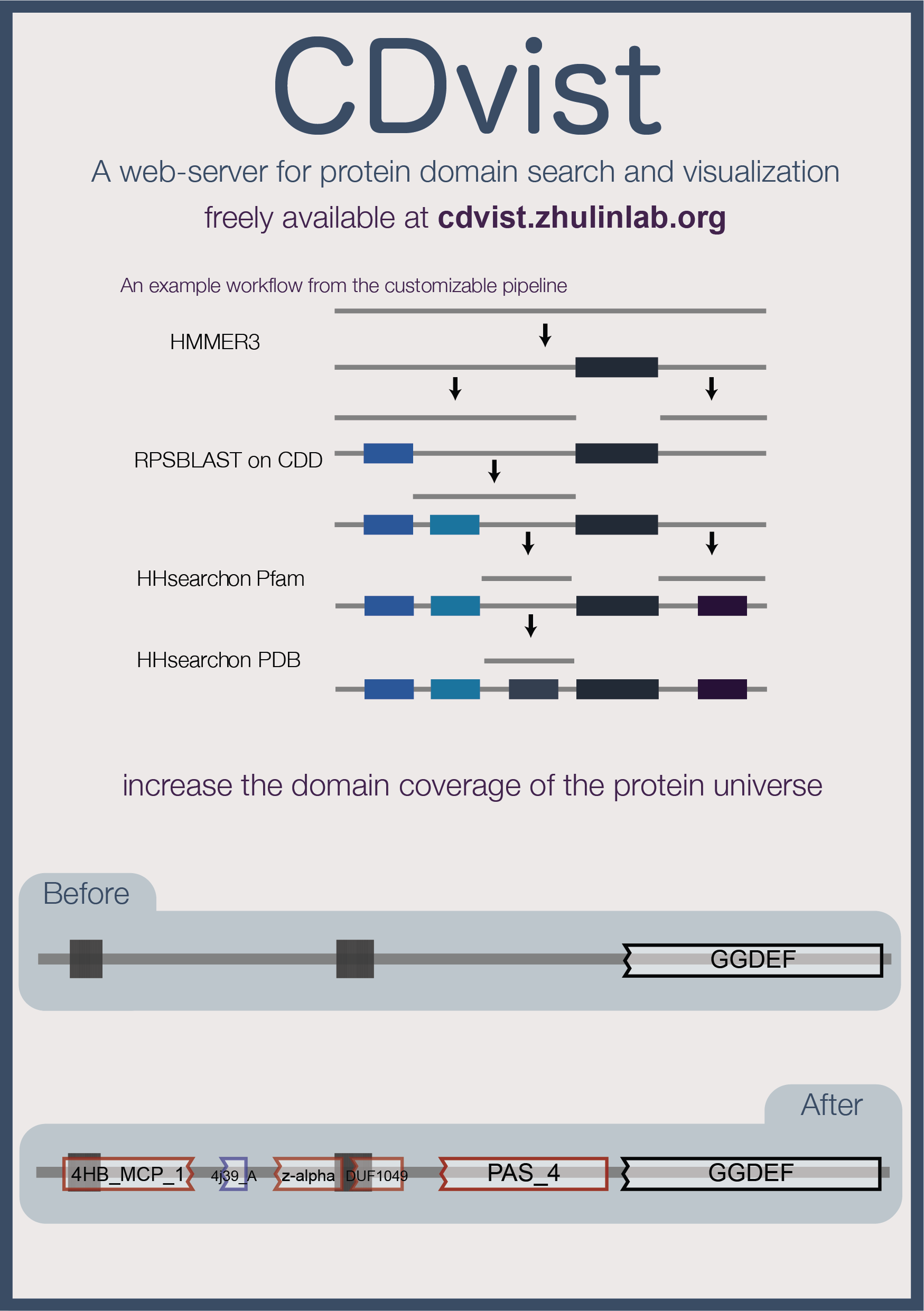
Identification of domains in protein sequences allows their assigning to biological functions. Several webservers exist for identification of protein domains using similarity searches against various databases of protein domain models. However, none of them provides comprehensive domain coverage while allowing bulk querying and their visualization schemes can be improved. To address these issues we developed CDvist (a comprehensive domain visualization tool), which combines the best available search algorithms and databases into a user-friendly framework. First, a given protein sequence is matched to domain models using high-specificity tools and only then unmatched segments are subjected to more sensitive algorithms resulting in a best possible comprehensive coverage. Bulk querying and rich visualization and download options provide improved functionality to domain architecture analysis.
- Related:
- Assessing predictions on fitness effects of missense variants in HMBS in CAGI6
- PHACTboost: A Phylogeny-aware Pathogenicity Predictor for the Missense Mutations via Boosting
- CAGI, the Critical Assessment of Genome Interpretation, establishes progress and prospects for computational genetic variant interpretation methods
- Aquerium: a web application for comparative exploration of domain-based protein occurrences on the taxonomically clustered genome tree.